KRAS mutation profile differences between rectosigmoid localized adenocarcinomas and colon adenocarcinomas
Introduction
Cancer is one of the major public health problems in the world. Globally, among common cancers, colorectal cancer is the fourth most common cancer in men and the third most common in women (1). Colorectal cancer has a heterogeneous nature that is influenced by the tumour site. Numerous studies have shown that risk factors, aetiology, clinical behaviour, and pathological and genetic features associated with colon and rectal cancers differ due to tumour location. Physical activity and healthy weight are associated with a reduced risk of colon cancer, but they are not associated with rectal cancer. Rectal tumours are more likely to be diagnosed in men than in women and in slightly younger patients compared to colon cancer patients. There are also some studies that have investigated carcinogenesis genes and their prognostic value in colon and rectal cancers. Nuclear β-catenin may have a different role in rectal cancers compared to colon cancers. The p53 pathway seems to be more significant in rectal cancers than in colon cancers. Many improvements have been made in identifying and characterizing the genetic alterations involved at the molecular level of colorectal carcinogenesis (2,3). The RAS/RAF/MAPK pathway is involved in cell proliferation and survival (4). V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) is mutated in 30-40% of sporadic colorectal cancers (5). However, there is insufficient information about KRAS mutation status and mutation type differences between colon and rectal cancers. KRAS mutations are useful markers for predicting responses to anti-EGFR monoclonal antibodies (moAbs) in metastatic colorectal cancers. Colorectal cancer patients with a KRAS mutation do not respond to treatment with cetuximab or panitumumab (6). KRAS mutations lead to constitutive activation of the RAS/RAF signalling pathway. This active signalling causes anti-EGFR monoclonal antibody-based treatment to be ineffective. Therefore, both the American Society of Clinical Oncology (ASCO) and the National Comprehensive Cancer Network (NCCN) recommend testing all metastatic colorectal cancer patients for KRAS mutations prior to anti-EGFR antibody treatment (7).
Surgical and therapeutic strategies may differ because of the variable nature of rectosigmoid cancer compared to colon cancer. The aim of this study was to compare the KRAS mutation test results of rectosigmoid cancers and colon cancers arising elsewhere in the large bowel with the results from proximal and distal colon cancers.
Methods
Tissue selection and DNA isolation
Eighty-four colorectal adenocarcinoma patients, whose archival tumour tissue was sufficient for molecular analysis, were chosen for this study between 2009-2011 years. KRAS mutation analysis was performed after a histological confirmation of cancer and the presence of >75% tumour cells in haematoxylin & eosin-stained slides by a pathologist.
Genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tumour tissue sections (10 μm thick) using a QIAamp DNA FFPE Tissue Kit (Qiagen, Cat no: 56404, Hilden, Germany), according to the manufacturer’s instructions. Three to five sections were used depending on the size of the tumour tissue sample. Tumour tissues were deparaffinised in xylene, washed with absolute ethanol and air dried. The lysis process was performed with proteinase K treatment at 56 °C overnight. The concentration and quality of the extracted DNA were determined by spectrometric measurement.
KRAS mutation analysis
The ARMS/Scorpion-based TheraScreen-KRAS Mutation Kit (Product code: KR-21, DxS Ltd, Manchester, UK) was used for KRAS mutation analysis. The assay is designed to detect the seven most common KRAS gene mutations (Gly12Ala, Gly12Asp, Gly12Arg, Gly12Cys, Gly12Ser, Gly12Val and Gly13Asp) on exon 2. Real-time PCR was performed on a real-time PCR platform (Lightcycler 480, Roche Applied Science, Mannheim, Germany) according to the recommendations of the manufacturer.
Statistical analysis
The SPSS (Statistical Package for the Social Sciences) (Version 19.0; SPSS, Inc., Chicago, IL, USA) program was used for statistical analysis. The following parameters were analyzed: age, gender, tumour location and KRAS gene molecular status. A Chi-square test was used to test the association between mutation status and other variables. A P value <0.05 was considered significant.
Ethics statement
The study was based on pathological archive material approved by Non-invasive Researches Clinical Research Ethics Committee (Approval date: 24.11.2011, protocol number: 373-GOA).
Results
Patient demographics
Eighty-four patients diagnosed with colorectal adenocarcinoma were included in this study. Among the 84 patients enrolled in this study, 35 (42%) patients were diagnosed with rectosigmoid cancer. Patient characteristics data are shown in Table 1. Among rectosigmoid cancers, 17 (48.6%) patients were male and 18 (51.4%) patients were female. The median age at diagnosis was 63.6 years (range, 48-87 years) for rectosigmoid cancers. Among the 49 (58%) colon cancer patients in this study, 31 (63.3%) of these patients were male and 18 (36.7%) patients were female. The median age at diagnosis was 62.22 years old (range, 42-81 years) for colon cancer patients. There were no significant correlations between KRAS mutations and age or gender in the rectosigmoid or colon tumour groups.
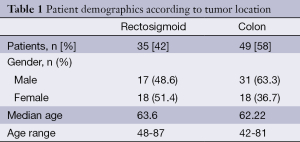
Full table
Molecular results
We analyzed the results of the KRAS mutation status in rectosigmoid-localized tumours versus colon-localized tumours. The distribution of KRAS mutations for rectosigmoid and colon tumours is shown in Table 2.
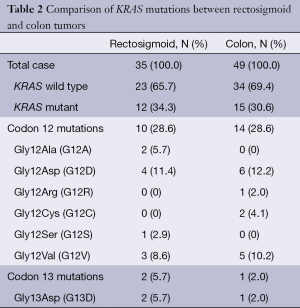
Full table
Rectosigmoid cancer cases: KRAS mutation analysis showed that 34.3% (12/35) of the rectosigmoid cancer patients had mutations at one of two codons (codon 12 and codon 13); 28.6% (10/35) of the rectosigmoid tumours had codon 12 mutations, and 5.7% (2/35) of the rectosigmoid tumours had codon 13 mutations. The mutation frequencies, according to their types, were determined as follows: G12D (11.4%, 4/35), G12V (8.6%, 3/35), G12A (5.7%, 2/35), G13D (5.7%, 2/35), and G12S (2.9%, 1/35). G12R and G12C mutations were not identified in rectosigmoid tumours. The KRAS mutation distributions according to gender are shown in Table 3.
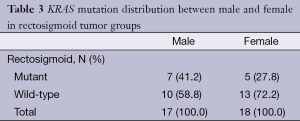
Full table
Colon cancer cases: mutation analysis showed that 30.6% (15/49) of the colon cancer patients had mutations at one of the codons; 28.6% (14/49) of the colon tumours had codon 12 mutations, and 2% (1/49) of the colon tumours had codon 13 mutations. The mutation frequencies, according to their types, were determined as follows: G12D (12.2%, 6/49), G12V (10.2%, 5/49), G12C (4.1%, 2/49), G12R (2%, 1/49), and G13D (2%, 1/49). G12A and G12S mutations were not identified in colon tumours. The KRAS mutation distributions according to gender are shown in Table 4.
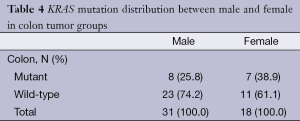
Full table
Conclusions
Colorectal cancers have a heterogeneous nature that is influenced by the tumour site. Several papers have explored the differences in clinicopathologic and/or molecular features between distal and proximal colon cancers. A higher frequency of allelic loss (17p, 18q), p53 mutations and c-myc expression in distal tumours has been reported previously. Higher microsatellite instability and more mucinous and diploid characteristics have been reported in proximal tumours (8-10). The clinical behaviour of rectal tumours is more variable than colon-localized tumours. Local recurrence is the major complication in rectal tumours, whereas distant metastasis is the most common issue in colon cancers. Some reviews have investigated the differences between rectal and colon tumours. The APC/beta-catenin and p53 pathways play a larger role in rectal cancers than in colon-localized cancers (2). Rectosigmoid cancers are classified as rectal tumours. The differences between rectosigmoid tumours compared to colon tumours have not been well studied. The current literature has generally focused on their surgical differences or on certain gene expression differences (11).
The molecular features of rectosigmoid and colon cancers may differ. KRAS mutations have functions in colorectal carcinogenesis and anti-EGFR therapy strategies. KRAS mutations lead to constitutively active RAS/RAF signalling. This constitutive activation causes anti-EGFR monoclonal antibody therapy to be ineffective in treating these tumours. We focused on KRAS gene mutation differences in rectosigmoid tumours. In the literature, there are no papers investigating KRAS mutations in rectosigmoid tumours. We present the KRAS genotypic features in rectosigmoid tumours and compare these results with colon-localized tumours.
In our study group, the rectosigmoid and colon group patients’ median age and ranges are similar. However, the gender distribution was not the same. The number of female patients is higher in the rectosigmoid tumour group, whereas in the colon tumour group, the number of male patients is higher. Some studies conducted on colorectal tumours have reported a higher frequency of KRAS mutations in females compared with males (12,13). In our study, male rectosigmoid cancer patients and female colon cancer patients had a higher frequency of KRAS mutations but no statistically significant difference between KRAS mutation and gender was observed in either rectosigmoid or colon tumours.
There was no significant correlation between KRAS mutations and tumour location in our rectosigmoid and colon cancer groups. In the literature, there were some correlations between KRAS mutation status and tumour location. However, these studies were based on colon, rectal or right/left-sided colon tumour groups and did not focus on rectosigmoid tumours. KRAS mutation frequency was found to be significantly higher in right-sided colon cancers in the study by Watanabe et al. Higher KRAS mutation frequency for right-sided colon cancers was presented in studies by Roth et al. and Abubaker et al. (14-16). In a study by Slattery et al., rectal and distal colon tumours were found to be more likely to carry KRAS mutations (3). In our group, the KRAS mutation frequency is higher in rectosigmoid tumours (34.3%, 12/35) than in colon localized tumours (30.6%, 15/49) (17). In rectal tumours, KRAS mutation rates are reported in 19% to 48% of patients. Slattery et al. reported KRAS mutation frequency in rectal tumours at 30.5% (3). The Catalogue of Somatic Mutations in Cancer (COSMIC) database which is designed to collect and present data regarding somatic mutations in cancer, shows that 35% of rectal adenocarcinomas carry a KRAS mutation (18). In our rectosigmoid group, the KRAS mutation frequency (34.3%) is similar to the study by Slattery et al. and the COSMIC database. In colon-localized tumours, KRAS mutation frequencies were reported at approximately 30%. These data are verified by the COSMIC database, which reports that 35% of colonic adenocarcinomas carry a KRAS mutation (18). In our colon group, the KRAS mutation frequency (30.6%) is similar to the COSMIC database.
The codon 12 and codon 13 mutation frequencies are similar between rectosigmoid and colon groups as 28.6% (10/35) of the rectosigmoid tumours had codon 12 mutations and 5.7% (2/35) of the rectosigmoid tumors had codon 13 mutations while 28.6% (14/49) of the colon tumours had codon 12 mutations and 2% (1/49) of the colon tumours had codon 13 mutations. The two most common mutations, G12D and G12V, were the same in both groups. Their frequencies were also similar. The most common mutations that we identified were the same as those of Chinese- and Serbian-based colorectal cancer patients (19,20). In rectosigmoid tumours, G12R and G12C mutations were not detected, while in colon tumours, G12A and G12S mutations were not detected. These mutations are the other most common mutations.
In summary, this study represents the first KRAS mutational results from Turkish rectosigmoid cancer patients. In this study, the KRAS mutation frequency in rectosigmoid tumours is higher than in colon localized tumours. KRAS mutations are involved in cell proliferation and survival. These mutations lead to constitutive activation of RAS/RAF signalling. This active signalling causes anti-EGFR monoclonal antibody-based treatments to be ineffective. KRAS mutation analysis has a predictive and prognostic value in identifying tumours that may be resistant to treatment. These results indicate that the aggressiveness of rectosigmoid tumours and their negative responses to therapy should be considered further. If this research is conducted with larger rectosigmoid and colon cancer groups, the comparison of the KRAS mutations may have significant correlations with other characteristics and all of the mutation types may be seen. Our study shows that the differences in biological behaviour between rectosigmoid and colon cancer should be considered. More studies are needed to understand the molecular behaviour of rectosigmoid cancer in our population. These results highlight the importance of personalized cancer management, which could be assisted by using cancer genotyping tools.
Acknowledgements
Funding: This work was supported by Dokuz Eylul University Research Foundation [Grant Number: DEU-BAP 2012.KB.SAG.63].
Disclosure: The authors declare no conflict of interest.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012;62:10-29. [PubMed]
- Kapiteijn E, Liefers GJ, Los LC, et al. Mechanisms of oncogenesis in colon versus rectal cancer. J Pathol 2001;195:171-8. [PubMed]
- Slattery ML, Curtin K, Wolff RK, et al. A comparison of colon and rectal somatic DNA alterations. Dis Colon Rectum 2009;52:1304-11. [PubMed]
- Russo A, Rizzo S, Bronte G, et al. The long and winding road to useful predictive factors for anti-EGFR therapy in metastatic colorectal carcinoma: the KRAS/BRAF pathway. Oncology 2009;77 Suppl 1:57-68. [PubMed]
- Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012;104:228-39. [PubMed]
- Yokota T. Are KRAS/BRAF mutations potent prognostic and/or predictive biomarkers in colorectal cancers? Anticancer Agents Med Chem 2012;12:163-71. [PubMed]
- Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 2009;27:2091-6. [PubMed]
- Fujita S, Baba H, Yamamoto S, et al. Allelic status of chromosomes 17p, 18q, 22q [corrected] 3p and their clinical usefulness in colorectal cancer. Anticancer Res 2006;26:2833-40. [PubMed]
- Soussi T. Advances in carcinogenesis: a historical perspective from observational studies to tumor genome sequencing and TP53 mutation spectrum analysis. Biochim Biophys Acta 2011;1816:199-208.
- Georgakopoulos G, Tsiambas E, Korkolopoulos P, et al. c-MYC and h-TERT co-expression in colon adenocarcinoma: a tissue microarray digitized image analysis. J BUON 2013;18:124-30. [PubMed]
- Birkenkamp-Demtroder K, Olesen SH, Sørensen FB, et al. Differential gene expression in colon cancer of the caecum versus the sigmoid and rectosigmoid. Gut 2005;54:374-84. [PubMed]
- Nagasaka T, Sasamoto H, Notohara K, et al. Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol 2004;22:4584-94. [PubMed]
- Patil H, Korde R, Kapat A. KRAS gene mutations in correlation with clinicopathological features of colorectal carcinomas in Indian patient cohort. Med Oncol 2013;30:617. [PubMed]
- Abubaker J, Bavi P, Al-Haqawi W, et al. Prognostic significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS mutations in colorectal carcinoma. J Pathol 2009;219:435-45. [PubMed]
- Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol 2010;28:466-74. [PubMed]
- Watanabe T, Yoshino T, Uetake H, et al. KRAS mutational status in Japanese patients with colorectal cancer: results from a nationwide, multicenter, cross-sectional study. Jpn J Clin Oncol 2013;43:706-12. [PubMed]
- Derbel O, Wang Q, Desseigne F, et al. Impact of KRAS, BRAF and PI3KCA mutations in rectal carcinomas treated with neoadjuvant radiochemotherapy and surgery. BMC Cancer 2013;13:200. [PubMed]
- Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 2011;39:D945-50. [PubMed]
- Jakovljevic K, Malisic E, Cavic M, et al. KRAS and BRAF mutations in Serbian patients with colorectal cancer. J BUON 2012;17:575-80. [PubMed]
- Mao C, Zhou J, Yang Z, et al. KRAS, BRAF and PIK3CA mutations and the loss of PTEN expression in Chinese patients with colorectal cancer. PLoS One 2012;7:e36653. [PubMed]


