Mismatch repair protein expression in colorectal cancer
Introduction
There are likely to be important clinical indications for determining the molecular subtypes of colorectal cancer. One parameter by which colorectal cancers can be classified involves the expression patterns of Mismatch repair (MMR) proteins. MMR proteins are nuclear enzymes, which participate in repair of base-base mismatch that occur during DNA replication in proliferating cells. The proteins form complexes (heterodimers) that bind to areas of abnormal DNA and initiates its removal. Loss of MMR proteins leads to an accumulation of DNA replication errors, particularly in areas of the genome with short repetitive nucleotide sequences, a phenomenon known as microsatellite instability (MSI) (1-3). MSI can be identified in more than 90% of colorectal cancers that arise in patients with Lynch syndrome, while in sporadic colorectal cancer it occurs in 15% of cases (4).
Mechanisms for MSI
Alterations in at least six of the genes that encode proteins involved in the MMR system have been identified in either hereditary nonpolyposis colorectal cancer (HNPCC) or sporadic colon cancer. These genes include MSH2, MSH3, MSH6, MLH1, PMS1, and PMS2. Study of the biochemistry of the MMR proteins has revealed that recognition of the base-base mismatches and insertion/deletion loops is performed by a heterodimer of either MSH2 and MSH6 or MSH2 and MSH3. Of interest, the MSH2-MSH3 heterodimer preferentially recognizes insertion/ deletion loops and thus cannot compensate for loss of MSH6. Consequently, cancers arising with a loss of MSH6 function display microsatellite instability only in mononucleotide repeats (5). The MLH1, PMS2, and PMS1 proteins appear to operate primarily in performing the repair of the base-base mismatches and insertion/deletion loops. A heterodimer of MLH1-PMS2 operates as a molecular matchmaker and is involved in executing the repair of the mismatches in conjunction with other molecules (5,6).
HNPCC related colon cancers account for 3-6% of all colon cancers, and germline mutations in MSH2 and MLH1 have been found in 45-70% of families that meet the Amsterdam criteria for HNPCC (7,8). Since inactivation of both alleles of MSH2 or MLH1 is required to generate MSI, the cancers that arise in HNPCC kindred frequently show loss of heterozygosity at the loci of these genes, or alternatively show somatic mutation of the sole wild-type MMR allele. The germline mutations that occur in MSH2 and MLH1 are widely distributed throughout either gene and are missense, deletion, or insertion mutations. These mutations result in frame shifts (60% of hMSH2 mutations and 40% of MLH1 mutations), premature truncations (23% of MSH2 mutations), or missense mutations (31% of MLH1 mutations) (9). The lack of a mutation hotspot has hampered the development of an inexpensive clinical assay to detect germline mutations in the genes known to cause HNPCC. Furthermore, because one wild-type allele is sufficient to maintain MMR activity, functional assays to detect MMR gene mutation carriers have not been developed for clinical use to date. However, proof-of-principle studies have demonstrated that it may be possible to develop such an assay by forcing a cell to a haploid state in which case a mutant MMR allele could be detected (10,11). Studies of the 15% of sporadic colon cancers that display MSI demonstrated these arose due to somatic inactivation of MMR genes and not due to germline MMR gene mutations with low penetrance. While occasional somatic mutations of MSH2 and MLH1 were detected, the predominant mechanism for inactivating MMR unexpectedly proved to be the epigenetic silencing of the MLH1 promoter due to aberrant promoter methylation (12,13).
Clinical implications of MSI
The CRC microsatellite profile provides useful prognostic information (14,15), showing the patients with microsatellite unstable neoplasms have a better overall survival rate and a modified response to conventional chemotherapy (16-21). MSI also helps in predicting the treatment response of CRC (18,19,22), and could modify the chemotherapy protocols offered to the patients in the future (19), but these results should be applied with caution before this predictive tool is verified.
Molecular markers as predictive factors in treatment decisions have been developed in the last few years. The initial studies in sporadic CRC showed that the retention of heterozygosity at one or more 17p or 18q alleles in microsatellite-stable CRCs and mutation of the gene for the type II receptor for TGF-β1 in CRCs with high levels of microsatellite instability correlated with a favorable outcome after adjuvant chemotherapy with fluorouracil based regimens, especially for stage III CRC (18,22). However, most recent studies have revealed that fluorouracil-based adjuvant chemotherapy benefited patients with stage II or stage III CRC with MSS tumors or tumors exhibiting low frequency MSI but not those with CRCs exhibiting high frequency MSI (19). The reasons for these responses must be related to the distinctive cell kinetics associated with MMR down-regulation (significantly increased apoptosis and decreased proliferation), which can certainly contribute to tumor cell resistance to conventional chemotherapy.
Testing for MSI and MMR defects
Clinical criteria
The recognition that certain types of cancers cluster in families with HNPCC and that cancer develops at relatively early ages compared with the general population provided the rationale for development of criteria that could be used to aid in the diagnosis. Two sets of criteria (the Amsterdam criteria and Bethesda guidelines) developed by a consensus of experts, have been most widely accepted and best studied.
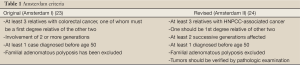
The Amsterdam criteria (Table 1) were designed to establish the diagnosis of HNPCC based upon familial clustering of HNPCC-related tumors. On the other hand, Bethesda guidelines (Table 2) were designed to help predict which patients with colorectal cancer are likely to have a mismatch-repair mutation and should thus undergo further testing. However, both the Amsterdam criteria and Bethesda guidelines have been studied for predicting the presence of mismatch repair mutations. Although the Bethesda guidelines and Amsterdam criteria continue to be used widely, several studies evaluating them (both the original and revised) have underscored the limitations of their accuracy in predicting the presence of mismatch repair mutations (25-28), and review of the literature reported that the sensitivity of the original Amsterdam criteria ranged from 54% to 91% (29). Such a wide range of estimates leaves substantial uncertainty as to the role of the Amsterdam criteria as a screening test for mismatch repair mutations. In addition to the limitations regarding their predictive accuracy, there are practical problems with policies based on the implementation of these clinical criteria. Patients’ report of the family history may not be accurate, particularly for cancers other than colorectal that are potentially related to HNPCC (30). Issues of uncertain paternity may also be relevant in some families while some families may be too small or have insufficient contact among family members to obtain a clinically meaningful family history.
Full Table
Clinical testing for MSI and MMR
Because of the limitations of relying on clinical criteria to guide testing, some authorities have proposed that tumors from patients with colorectal cancer be evaluated for markers of HNPCC regardless of the family history (23,24). One of the largest studies evaluating this approach included 1066 patients with colorectal cancer whose tumors were tested for MSI (23). Patients with suggestive MSI results were tested for germ-line mutations in the mismatch repair genes (MSH2, MLH1, MSH6, and PMS2) by IHC, genomic sequencing, and deletion studies. A mutation causing HNPCC was detected in 23 patients (2.2 percent) of whom ten were older than 50 and five did not meet the Amsterdam criteria or Bethesda guidelines. These data suggest that the Amsterdam or Bethesda criteria alone may miss as many as 22 percent of patients with HNPCC. However, only five additional individuals from the cohort of 1,066 subjects (0.5%) would have been identified by routine molecular analysis of all colon cancers fulfilling the Bethesda criteria, making such an approach impractically expensive for routine clinical use. Therefore; most expert guidelines on HNPCC suggest a combination of sequential laboratory testing in patients who fulfill the Amsterdam criteria or Bethesda guidelines to minimize costs and maximize test accuracy (31,32). Approaches based on such a strategy have been considered to be cost-effective (33). However, the exact methods and order of testing are unsettled. Proposed strategies include initial testing of tumors for MSI with or without IHC for loss or expression of mismatch repair proteins, with germline gene sequencing reserved for patients with suggestive results.
Microsatellite instability (MSI) testing
MSI testing involves amplification of a standardized panel of DNA markers; five markers were agreed upon by a consensus panel convened by the National Institutes of Health in 1997 (15). The reference panel included two mononucleotide markers (BAT25 and BAT26) and three dinucleotide microsatellites (D5S346, D2S123 and D17S250), previously tested by Fishel (34), plus a list of several alternative loci. Three categories of MSI have been recognized based upon these panels: MSI-high (instability of two or more markers), MSI-low (instability of one marker), and MS-stable (no instability). More recently, some laboratories have begun using ten or more markers. In such cases MSI is defined as stable when fewer than 10% of markers are unstable, low when 10 to 30% of markers are unstable and high when greater than 30-40% of markers are unstable. There are several pitfalls of MSI testing. First, it is labor intensive, relatively costly, and requires expert pathologic services. In addition, tissue to be amplified should ideally be microdissected to avoid amplifying DNA from normal colonic mucosa.
Immunohistochemistry (IHC) testing
Pathogenic mutations in MMR proteins usually lead to the absence of a detectable gene product providing the rational for immunohistochemistry testing to determine loss of expression. Tumours from patients suspected to have MSI can be stained for MMR proteins and the surrounding normal tissues can be used as a positive control. IHC has an advantage over MSI analysis as it is much easier to perform and less expensive. Moreover, it provides gene specific information to direct further genetic analysis. However; the technique is vulnerable to the quality of tissue preparation, staining and interpretation.
The understanding of how the MMR proteins interact during DNA repair can help in the interpretation of the results of such testing. MSH2 forms a heterodimer with MSH6, while MLH1 binds to PMS2 and complexes MSH2/MSH6 heterodimer. Therefore, when MSH6 is not detected in a tumour MSH6 may also not detected. The situation is more complex with lack of MLH1 expression. Hypermethylation of hMLH1 gene, which is common in sporadic colorectal cancer, may lead to loss of protein expression.
IHC has a role in detecting MMR defects, with data suggesting that the effectiveness of IHC screening of the MMR proteins would be similar to that of the more complex strategy of microsatellite genotyping (23,25). This technique can guide which gene to sequence and can help differentiating sporadic from hereditary mutations: MSH2 loss is likely to be HNPCC, whereas MLH1 loss could be HNPCC or sporadic CRC (MLH1 promoter methylation). MMR proteins heterodimerize to function; the MSH2 loss almost always accompanies MSH6 loss and when MLH1 is lost, generally so is hPMS2 (35,36). In addition, IHC can miss functional loss; i.e. presence of the protein with antigen positivity in the absence of function.
MMR IHC studies are based on a complete absence of at least one MMR protein (37-41). But these studies do not consider the immunostaining topographic heterogeneity. Since the MMR proteins function as heterodimers, it could be advocated to validate the IHC results of MSH2/MSH6 and MLH1/PMS2. More studies are required to clarify the influence of this predictable tumor heterogeneity to select the appropriate sample for immunohistochemical and/or MSI analyses
Genetic testing
Multiple methods have been used for genetic testing in HNPCC. The methods used should ideally be able to detect the many potential genotypes associated with HNPCC like nonsense, missense, and frame shift mutations, genomic deletions, duplications, and rearrangements. The commonly used tests includes: high output screening techniques, DNA sequencing, conversion analysis and methods to detect large structural DNA abnormalities like Southern blot and Multiplex ligation-dependent probe amplification.
Aims
Information about MMR protein status in colorectal cancer is important because it will identify those most likely to have Lynch syndrome and those most likely to have microsatellite instability in their tumours which has been proven to have better prognosis and may affect their treatment regimens in the future. We undertook this study to develop and optimise a protocol for MMR protein immunohistochemistry testing in colorectal cancer. We also aimed to analyse the proportion of patients with colorectal cancer with loss of immunostaining for MMR proteins (hMLH1, hMPS2, hMSH2 and hMSH6) in order to determine the feasibility of molecular screening for the loss of MMR proteins through the study of unselected patients with colorectal cancer.
Materials and methods
Study group
A group of 33 patients with colorectal cancer was randomly selected from the department of surgery bio-bank to determine the expression of MMR proteins in their FFPE tumour tissues using immunohistochemistry techniques. The age of the patients at diagnosis of their cancers and their family history were collected by reviewing the medical charts.
FFPE tissues
Tumour tissues collected at time of surgery were collected and placed in 10% formalin (Lennox) for fixation at room temperature until embedding for a minimum of 24 hours. Tissue was then removed from the formalin and placed on an open cassette. The cassette was closed and placed in 250 mL of Industrial Methylated Spirit (VWR) to wash the formalin from the tissue. Then, the cassette was removed and placed in JFC solution (Milestone) filed JFC beaker and placed in the histoprocessor (MicroMED) for 60 minutes (70 °C). Thereafter, the cassette was transferred to the paraffin wax (VWR) filled wax beaker and placed in the histoprocessor (MicroMED) for 30 minutes. The cassette was removed from the wax beaker and tissue was blocked out carefully. The blocks were left at 4 °C until hard and then stored at fridge or room temperature until sectioning. Sectioning of formalin-fixed paraffin-embedded tissues was carried out using Slee microtome (LIS Ltd). With section thickness set to 30µM the block was pared down until even sections were being cut and the outer layer of wax was removed. Then the section thickness was adjusted to 5 µM. The sections were then placed in a floating out bath to stretch it out, before being placed onto a Superfrost plus (positive charged) slides (VWR). The slides were allowed to air-dry overnight at room temperature and then stored at 4 °C until further use. Before enrolment in any further experiments each slide is stained in H & E and reviewed by a pathologist to determine the quality of the block and the percentage of tumour tissues in the section (should be >50%)
Immunohistochemistry
Immunostaining was carried out on 5 µm thick paraffin sections of tumour tissue from each patient, using mouse monoclonal antibodies specific for each of the four human MMR proteins and employing automated DABMap system (Ventana) for hMSH6 detection and UltraMap system (Ventana) to detect hMLH1, hMSH2, and hPMS2 proteins.
DABMap protocol
It was consist of deparaffinization and cell conditioning, followed by addition of primary antibody and incubation at room temperature for I hour. Then the secondary antibody was added before counterstaining with haematoxylin and slides dehydration.
UltraMap protocol
The standard UltraMap was used to detect hMSH2. It was again consist of deparrafinization and cell conditioning followed by primary antibody titration. The tissue section was incubated with primary antibody for 12 hours at 37 °C. No secondary antibody was added. This was followed by counterstaining and dehydration in serial ethanol alcohol dilution and Xylene (Sigma).
The extended UltraMap protocol was used to determine the expression of hMLH1 and hPMS2. It was different from the standard one in that the cell conditioning was extended to three cycles of medium cell conditioner and cell conditioner compared to two cycles in case standard protocol.
IHC analysis
Changes in protein expression following transfection of colorectal tissues were observed in stained cells using Olympus BX60 microscope and image analySIS software. Adjacent normal tissue served as an internal control for positive staining and a negative control staining was carried out without the primary antibody. MMR protein staining was considered negative when all of the tumour cell nuclei failed to react with the antibody.
Results
Optimization of MMR protein staining protocol
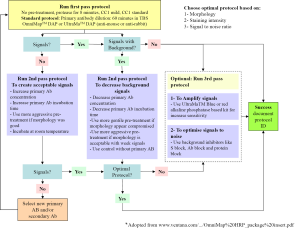
Tissue processing has the greatest single impact on the end result of IHC and different tissue types often require slightly different pre-treatments for optimum results. To optimized staining protocols we employed the Closed Loop Assay Development (CLAD) for IHC (Figure 1).
Optimal staining was achieved for hMSH6 using DABMap system, however; acceptable stating for hMLH1, hMSH2 and hMPS2 was only achievable using UltarMap system.
MMR protein expression
IHC staining was performed on 33 colorectal cancer tissue specimens. Loss of MMR protein is defined as complete absence of nuclear staining within the tumour. While MMR proteins expression is defined as the presence of nuclear staining within the tumour regardless its intensity or the number of positive nuclei (Figures 2-5)
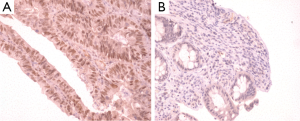
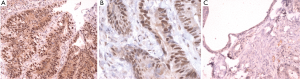
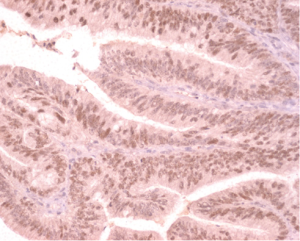
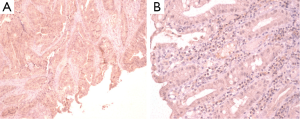
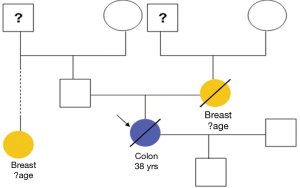
Of the tissue specimens in which acceptable immunostaining was achieved, three samples showed loss of one or more of the MMR proteins (Table 3). Both hMLH1 and hPMS2 proteins were not expressed in a 36 years old woman (case 3) with cancer of the caecum (Proximal to the splenic flexure). She had history of breast cancer on her mother and colorectal cancer on one of her grandfathers (undocumented weather on paternal or maternal side) (Figure 6). The expression of hMSH6 protein was undetermined in tumour tissues retrieved from a 61 years old man (case 13) with cancer of the proximal colon (proximal to the splenic flexure). He had no documented family history of cancer. The third case was a 77 years old man (case 27), again with no documented family history of cancer, who had carcinoma of the rectum. He showed loss of hMLH1 expression in the tumour tissues.
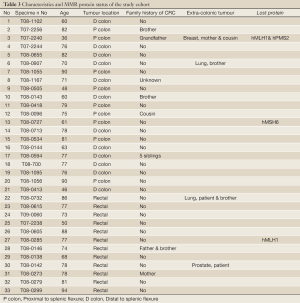
Full Table
Discussion
The identification of HNPCC can be lifesaving as it can lead to early detection of cancer. Jarvinen et al. in a controlled clinical trial extending over 15 year period concluded that screening for colorectal cancer in HNPCC families more than halves the risk of colorectal cancer, prevents deaths from colorectal cancer and decreases the overall mortality rate by about 65% (42). Furthermore; the cost-effectiveness of screening was quantified by Ramsey et al. as $7,556 per year of life gained (33).When clinical and pedigree criteria such as Amsterdam criteria are used to determine what proportion of all colorectal cancers are due to HNPCC, estimate range from 1-6% (43). However; molecular screening has suggested that more 3% of all such patients have HNPCC. Moreover, the mean age at presentation with HNPCC diagnosed by molecular screening was 54 years old in a study included several patients over 60 years of age (44,45).
In addition, experiments have recently shown the differences in the response of MSI-H tumours to chemotherapeutic agents. DNA mismatched repair-deficient cells are resistant to the alkylating agents (e.g., melphalan and busulphan), methylating agents (e.g., temozolomide), the platinum-containing agents (e.g., cisplatin and carboplatin), antimetabolites (e.g. fluorouracil and thioguanine) and topoisomerase inhibitors (e.g., doxorubicin) (46,47). The clinical significance of these observations remained unclear till recently. A meta-analysis of 32 studies with 7,642 cases found the hazard ratio (HR) for overall survival in patients whose tumours have high microsatellite instability (MSI-H) is 0.65 (95% CI: 0.59-0.71). Two studies, in this review, have assessed the benefit of 5-fluorouracil (5-FU) in stage II and III colorectal cancer patients by MSI status. The analysed data indicates that patients without MSI benefited significantly from 5-FU (HR=0.72, 95% CI: 0.61-0.84), while patients with MSI did not benefit from 5-FU (HR=1.24, 95% CI: 0.72-2.14) (48).
Because of the limitations of relying on clinical criteria to guide testing for Lynch syndrome and the prognostic information that could be provided by MSI status, molecular screening of all patients with colorectal cancer for MMR protein expression is now both feasible and desirable. In most Lynch syndrome colorectal tumours, MSI has been shown to result from defects in DNA mismatch repair mechanism (49). Mutations in hMLH1 or hMSH2 genes are the most common defects in these families making up to 94% of the germ line mutations detected. In addition, a few families have been found to have hMSH6 or hPMS2 mutations (9,50). On the other hand, about 10-15% of sporadic colorectal cancer also exhibit MSI, and loss of one or more of the MMR proteins has been found in these tumours (14,51). Lack of expression of hMLH1 as the result of promoter methylation occurs in most of sporadic MSI-positive tumours (52). Loss of the other MMR proteins is rare in sporadic tumours and in one study loss of either hMSH2 or hPMS2 was found in only 2% of tumours (53).
The major laboratory tests used in the evaluation of patients suspected to have Lynch syndrome include testing of tumour tissues using immunohistochemistry (IHC), MSI testing or germ line testing for mismatch defects. IHC has the advantage over the other methods, as the primary screening method, since it is less demanding to perform and is available as part of routine services in general pathology laboratories. In addition, IHC will determine which protein is affected and provides gene specific information; thereby direct the genetic analysis rather than performing exhausting, time and material consuming unnecessary tests. Nevertheless, while most of mutations will results in total loss of the protein expression, in some cases mutations only result in loss of function rather than the expression of the protein which will still be detectable by IHC.
Many studies have provided information about the sensitivity and specificity of IHC for predicting MMR mutations (25,37,54-60). A recent meta-analysis determined the sensitivity to range from 27%-100% and specificity from 43-100%, however, analysis of good quality studies only had a summary sensitivity of 74% (95% CI: 54-87) and specificity of 77% (95% CI: 61-88) (61). In one study of unselected 131 colorectal cancer patients diagnosed younger than 45 years of age the sensitivity of IHC testing for the main 4 MMR proteins was reported as 100% and its specificity was 69% (60). Lindoe et al. have assessed 1,144 patients with colorectal cancer for MMR deficiency by MSI testing and IHC detection for hMLH1 and hMSH2. They determined 92% specificity and 100% specificity of IHC for screening for MMR defects (62).
In evaluating the expression of MMR proteins using IHC, any tumour cell nuclear expression is considered positive due to the heterogeneity of expression and difficulties in test standardisation (63). The intensity of staining in normal mucosa decreased towards the surface. Moreover, the normal enterocytes can serve as positive internal controls and should always be observed to determine the quality of staining (64). In sporadic tumours due to hypermethylation of the promoter of hMLH1 there is consistent loss of the protein expression (65). Therefore, this feature alone can not differentiate sporadic MSI-H tumours from Lynch syndrome due to germLine mutation in hMLH1 (approximately half of the cases) and methylation analysis would more help in the determination of the nature of mutation.
In this study, we looked the MMR protein expression without considering the family history or the result of previous tumour testing for microsatellite status in a prospective of newly diagnosed colorectal cancer patients. We identified three patients with loss of one or more MMR protein. The first patient (case 3) was less than 40 years old when diagnosed with caecal cancer. Although her family history was not fully documented (Figure 6), she showed history of colorectal and breast cancer in some members of her family. Her tumour loss the expression of hMLH1 and hPMS2, making her more likely to have Lynch syndrome. The other two cases were more than 60 years of age when diagnosed with colorectal cancer which is not a typical age for tumour onset in Lynch syndrome patients. However; case 13 who loss the expression of hMSH6 in his proximal colon tumour can still have Lynch syndrome. Case 27 was 77 years old when developed a rectal cancer. The loss of hMLH1 expression in his tumour in addition to the lack of family history of cancer makes him more likely to have microsatellite instable sporadic cancer. Our results are in keeping with previous report by Hamplel et al. (23). They examined 1,066 patients with newly diagnosed colorectal adenocarcinoma for MSI. Among patients whose screening results were positive for MSI, they looked for germLine mutations in the 4 main MMR genes using IHC, genomic sequencing and deletion studies. MSI was detected in 19.5% of their study population and 2.2% were confirmed to have Lynch syndrome. Of the patients who were found to have Lynch syndrome 10 were more than 50 years and 5 did not meet the clinical criteria for diagnosis of HNPCC. Their data suggested the similar efficiency of IHC and the more complex genetic analysis for MSI testing.
Our findings and the previous reports pointed out the importance of molecular screening of patients with colorectal cancer for MSI using immunohistochemistry. This strategy managed to identify mutations in patients otherwise would not have been detected. Therefore, we recommend it as a policy for all newly diagnosed colorectal cancer patients due to its important prognostic implications.
Acknowledgements
We would like to thank the National Breast Cancer Research Institute (NBCRI) for their financial support of the study
Disclosure: The authors declare no conflict of interest.
References
- Ionov Y, Peinado MA, Malkhosyan S, et al. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 1993;363:558-61. [PubMed]
- Aaltonen LA, Peltomäki P, Leach FS, et al. Clues to the pathogenesis of familial colorectal cancer. Science 1993;260:812-6. [PubMed]
- Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science 1993;260:816-9. [PubMed]
- Aaltonen LA, Peltomäki P, Mecklin JP, et al. Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal cancer patients. Cancer Res 1994;54:1645-8. [PubMed]
- Jiricny J. Replication errors: cha(lle)nging the genome. EMBO J 1998;17:6427-36. [PubMed]
- Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev 1999;9:89-96. [PubMed]
- Marra G, Boland CR. Hereditary nonpolyposis colorectal cancer: the syndrome, the genes, and historical perspectives. J Natl Cancer Inst 1995;87:1114-25. [PubMed]
- Liu B, Parsons R, Papadopoulos N, et al. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med 1996;2:169-74. [PubMed]
- Peltomäki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 1997;113:1146-58. [PubMed]
- Yan H, Papadopoulos N, Marra G, et al. Conversion of diploidy to haploidy. Nature 2000;403:723-4. [PubMed]
- Langer S, Jentsch I, Gangnus R, et al. Facilitating haplotype analysis by fully automated analysis of all chromosomes in human-mouse hybrid cell lines. Cytogenet Cell Genet 2001;93:11-5. [PubMed]
- Kane MF, Loda M, Gaida GM, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 1997;57:808-11. [PubMed]
- Veigl ML, Kasturi L, Olechnowicz J, et al. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci U S A 1998;95:8698-702. [PubMed]
- Lawes DA, SenGupta S, Boulos PB. The clinical importance and prognostic implications of microsatellite instability in sporadic cancer. Eur J Surg Oncol 2003;29:201-12. [PubMed]
- Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248-57. [PubMed]
- Elsaleh H, Powell B, Soontrapornchai P, et al. p53 gene mutation, microsatellite instability and adjuvant chemotherapy: impact on survival of 388 patients with Dukes' C colon carcinoma. Oncology 2000;58:52-9. [PubMed]
- Barratt PL, Seymour MT, Phillips RM, et al. The role of the DNA mismatch repair protein, hMLH1 in sensitivity of colon colon cancer cells to chemotherapeutic agents used clinically. Br J Cancer 1998;78:9.
- Barratt PL, Seymour MT, Stenning SP, et al. DNA markers predicting benefit from adjuvant fluorouracil in patients with colon cancer: a molecular study. Lancet 2002;360:1381-91. [PubMed]
- Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 2003;349:247-57. [PubMed]
- Elsaleh H, Powell B, McCaul K, et al. P53 alteration and microsatellite instability have predictive value for survival benefit from chemotherapy in stage III colorectal carcinoma. Clin Cancer Res 2001;7:1343-9. [PubMed]
- Chen X, Lai MD, Huang Q. Increased sensitivity of colorectal cancer cell lines with microsatellite instability to 5-fluorouracil in vitro. Chin Med J (Engl) 2002;115:1048-52. [PubMed]
- Watanabe T, Wu TT, Catalano PJ, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med 2001;344:1196-206. [PubMed]
- Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (Hereditary nonpolyposis colorectal cancer). N Engl J Med 2005;352:1851-60. [PubMed]
- Ward RL, Turner J, Williams R, et al. Routine testing for mismatch repair deficiency in sporadic colorectal cancer is justified. J Pathol 2005;207:377-84. [PubMed]
- Barnetson RA, Tenesa A, Farrington SM, et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med 2006;354:2751-63. [PubMed]
- Piñol V, Castells A, Andreu M, et al. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA 2005;293:1986-94. [PubMed]
- Wüllenweber HP, Sutter C, Autschbach F, et al. Evaluation of Bethesda guidelines in relation to microsatellite instability. Dis Colon Rectum 2001;44:1281-9. [PubMed]
- Raedle J, Trojan J, Brieger A, et al. Bethesda guidelines: relation to microsatellite instability and MLH1 promoter methylation in patients with colorectal cancer. Ann Intern Med 2001;135:566-76. [PubMed]
- Gruber SB. New developments in Lynch syndrome (hereditary nonpolyposis colorectal cancer) and mismatch repair gene testing. Gastroenterology 2006;130:577-87. [PubMed]
- Murff HJ, Spigel DR, Syngal S. Does this patient have a family history of cancer? An evidence-based analysis of the accuracy of family cancer history. JAMA 2004;292:1480-9. [PubMed]
- Giardiello FM, Brensinger JD, Petersen GM. AGA technical review on hereditary colorectal cancer and genetic testing. Gastroenterology 2001;121:198-213. [PubMed]
- Chung DC, Rustgi AK. The hereditary nonpolyposis colorectal cancer syndrome: Genetics and clinical implications. Ann Intern Med 2003;138:560-70. [PubMed]
- Ramsey SD, Clarke L, Etzioni R, et al. Cost-effectiveness of microsatellite instability screening as a method for detecting hereditary nonpolyposis colorectal cancer. Ann Intern Med 2001;135:577-88. [PubMed]
- Dietmaier W, Wallinger S, Bocker T, et al. Diagnostic microsatellite instability: Definition and correlation with mismatch repair protein expression. Cancer Res 1997;57:4749-56. [PubMed]
- Drummond JT, Li GM, Longley MJ, et al. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science 1995;268:1909-12. [PubMed]
- Prolla TA, Pang Q, Alani E, et al. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science 1994;265:1091-3. [PubMed]
- Wahlberg SS, Schmeits J, Thomas G, et al. Evaluation of microsatellite instability and inummohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res 2002;62:3485-92. [PubMed]
- Hawkins NJ, Ward RL. Sporadic colorectal cancers with microsatellite instability and their possible origin in hyperplastic polyps and serrated adenomas. J Natl Cancer Inst 2001;93:1307-13. [PubMed]
- Parc YR, Halling KC, Wang L, et al. HMSH6 alterations in patients with microsatellite instability-low colorectal cancer. Cancer Res 2000;60:2225-31. [PubMed]
- Alexander J, Watanabe T, Wu TT, et al. Histopathological identification of colon cancer with microsatellite instability. Am J Pathol 2001;158:527-35. [PubMed]
- Thibodeau SN, French AJ, Cunningham JM, et al. Microsatellite instability in colorectal cancer: different mutator phenotypes and the principal involvement of hMLH1. Cancer Res 1998;58:1713-8. [PubMed]
- Järvinen HJ, Aarnio M, Mustonen H, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 2000;118:829-34. [PubMed]
- Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med 2003;348:919-32. [PubMed]
- Aaltonen LA, Salovaara R, Kristo P, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med 1998;338:1481-7. [PubMed]
- Salovaara R, Loukola A, Kristo P, et al. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol 2000;18:2193-200. [PubMed]
- Fink D, Aebi S, Howell SB. The role of DNA mismatch repair in drug resistance. Clin Cancer Res 1998;4:1-6. [PubMed]
- Anthoney DA, McIlwrath AJ, Gallagher WM, et al. Microsatellite instability, apoptosis, and loss of p53 function in drug-resistant tumor cells. Cancer Res 1996;56:1374-81. [PubMed]
- Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 2005;23:609-18. [PubMed]
- Peltomäki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet 2001;10:735-40. [PubMed]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell 1996;87:159-70. [PubMed]
- Chaves P, Cruz C, Lage P, et al. Immunohistochemical detection of mismatch repair gene proteins as a useful tool for the identification of colorectal carcinoma with the mutator phenotype. J Pathol 2000;191:355-60. [PubMed]
- Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A 1998;95:6870-5. [PubMed]
- Plaschke J, Krüger S, Pistorius S, et al. Involvement of hMSH6 in the development of hereditary and sporadic colorectal cancer revealed by immunostaining is based on germline mutations, but rarely on somatic inactivation. Int J Cancer 2002;97:643-8. [PubMed]
- Curia MC, Palmirotta R, Aceto G, et al. Unbalanced germ-line expression of hMLH1 and hMSH2 alleles in hereditary nonpolyposis colorectal cancer. Cancer Res 1999;59:3570-5. [PubMed]
- Debniak T, Kurzawski G, Gorski B, et al. Value of pedigree/clinical data, immunohistochemistry and microsatellite instability analyses in reducing the cost of determining hMLH1 and hMSH2 gene mutations in patients with colorectal cancer. Eur J Cancer 2000;36:49-54. [PubMed]
- Dieumegard B, Grandjouan S, Sabourin JC, et al. Extensive molecular screening for hereditary non-polyposis colorectal cancer. Br J Cancer 2000;82:871-80. [PubMed]
- Syngal S, Fox EA, Eng C, et al. Sensitivity and specificity of clinical criteria for hereditary non-polyposis colorectal cancer associated mutations in MSH2 and MLH1. J Med Genet 2000;37:641-5. [PubMed]
- Terdiman JP, Gum JR Jr, Conrad PG, et al. Efficient detection of hereditary nonpolyposis colorectal cancer gene carriers by screening for tumor microsatellite instability before germline genetic testing. Gastroenterology 2001;120:21-30. [PubMed]
- Durno C, Aronson M, Bapat B, et al. Family history and molecular features of children, adolescents, and young adults with colorectal carcinoma. Gut 2005;54:1146-50. [PubMed]
- Southey MC, Jenkins MA, Mead L, et al. Use of molecular tumor characteristics to prioritize mismatch repair gene testing in early-onset colorectal cancer. J Clin Oncol 2005;23:6524-32. [PubMed]
- Bonis PA, Trikalinos TA, Chung M, et al. Hereditary Nonpolyposis Colorectal Cancer: Diagnostic Strategies and Their Implications, in Evidence Report/Technology. 2007, Tufts-New England Medical Center Evidence-based Practice Center: Rockville, MD: Agency for Healthcare Research and Quality.
- Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol 2002;20:1043-8. [PubMed]
- Gatalica Z, Torlakovic E. Pathology of the hereditary colorectal carcinoma. Fam Cancer 2008;7:15-26. [PubMed]
- Müller W, Burgart LJ, Krause-Paulus R, et al. The reliability of immunohistochemistry as a prescreening method for the diagnosis of hereditary nonpolyposis colorectal cancer (HNPCC)--results of an international collaborative study. Fam Cancer 2001;1:87-92. [PubMed]
- Young J, Simms LA, Biden KG, et al. Features of colorectal cancers with high-level microsatellite instability occurring in familial and sporadic settings: parallel pathways of tumorigenesis. Am J Pathol 2001;159:2107-16. [PubMed]